Layout Embedding#
minorminer.layout.find_embedding() offers a more specialized approach to
find an embedding through the use of the Layout
and Placement classes. This kind of
embedding may be useful when the underlying data of your source graph is spatial. It can also be useful for
embedding graphs with nodes of a low degree (i.e., a cubic graph).
- find_embedding(S, T, layout=(<function p_norm>, None), placement=<function closest>, mm_hint_type='initial_chains', return_layouts=False, **kwargs)[source]#
Tries to embed S in T by computing layout-aware chains and passing them to
minorminer.find_embedding(). Chains are passed as either initial_chains or suspend_chains (see documentation forminorminer.find_embedding()to learn more).- Parameters:
S (NetworkX Graph/edges data structure (dict, list, ...)) – The source graph being embedded or a NetworkX supported data structure for edges (see
nx.convert.to_networkx_graph()for details).T (NetworkX Graph/edges data structure (dict, list, ...)) – The target graph being embedded into or a NetworkX supported data structure for edges (see
nx.convert.to_networkx_graph()for details).layout (function/(function/dict/Layout, function/dict/Layout), optional) –
A function to compute the
Layoutfor both S and T, or a 2-tuple that either consists of a pair of functions or pre-computed layouts (in the form ofLayoutor dicts). The first entry in the 2-tuple applies to S while the second applies to T.Note
If
layoutis a single function and T is a dnx_graph, then the function passed in is only applied to S and the dnx_layout is applied to T. To run a layout function explicitly on T, pass it in as a 2-tuple; i.e. (p_norm, p_norm).placement (function/dict, optional, default=minorminer.placement.closest) –
A function that uses the layouts of S and T to map the vertices of S to subsets of vertices of T (
Placement), or a dict that contains the precomputed mapping/Placement.By default,
closest()is called to compute placement.mm_hint_type (str, optional, default="initial_chains") – This is the hint type passed to
minorminer.find_embedding(). Supported types are “initial_chains” and “suspend_chains”. Seeminorminer.find_embedding()for more information.return_layouts (bool, optional, default=False) – If True, layout objects of S and T are also returned.
**kwargs (dict) – Keyword arguments passed to
Layout,Placementorminorminer.find_embedding().
- Returns:
An embedding of vertices of S (keys) to chains in T (values). This embedding is dependent on the kwargs being passed in. If
return_layoutsis True, a 2-tuple is returned in which the first element is the embedding dict and the second element is another 2-tuple containing the source and targetLayoutobjects.- Return type:
Examples#
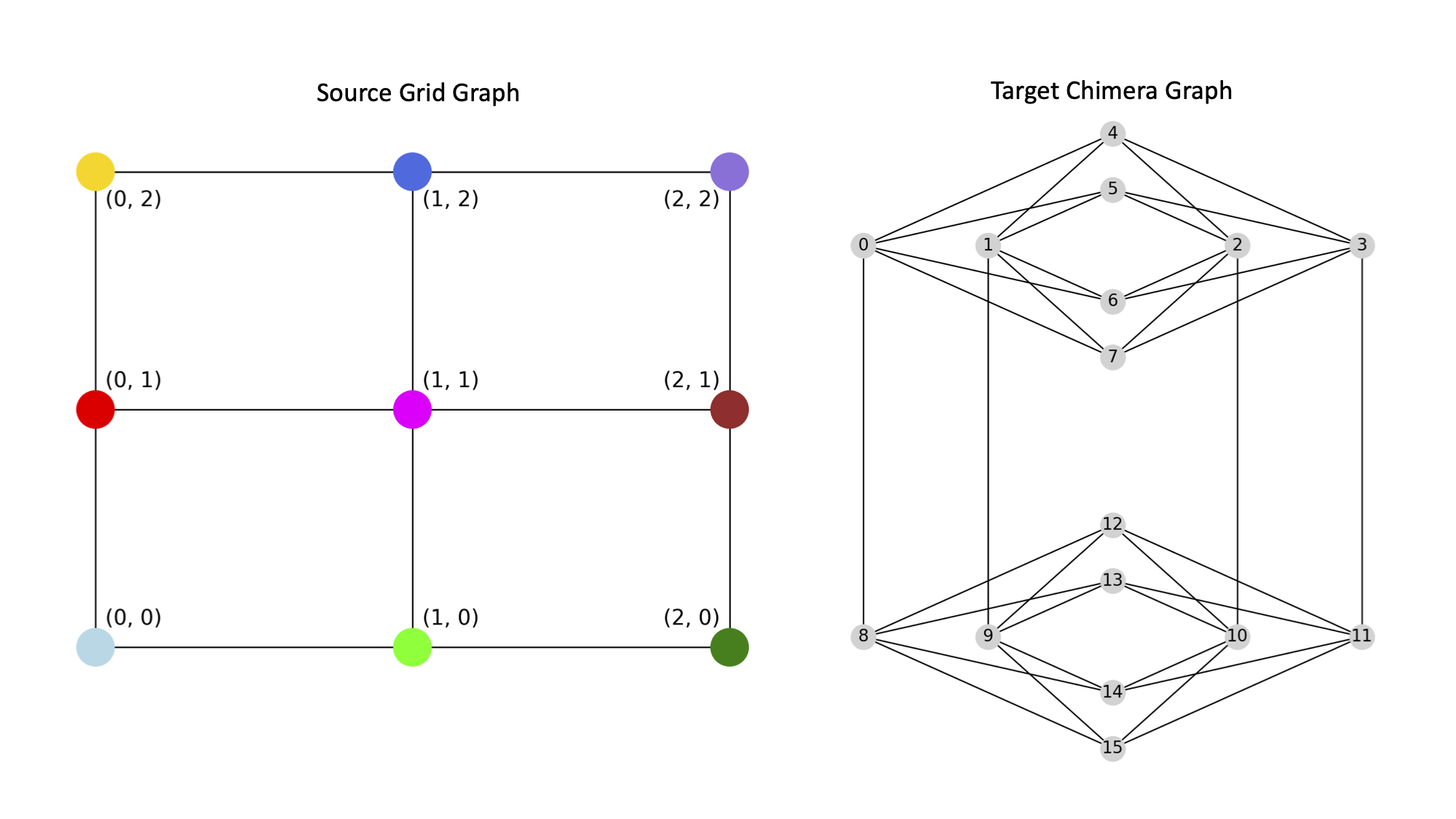
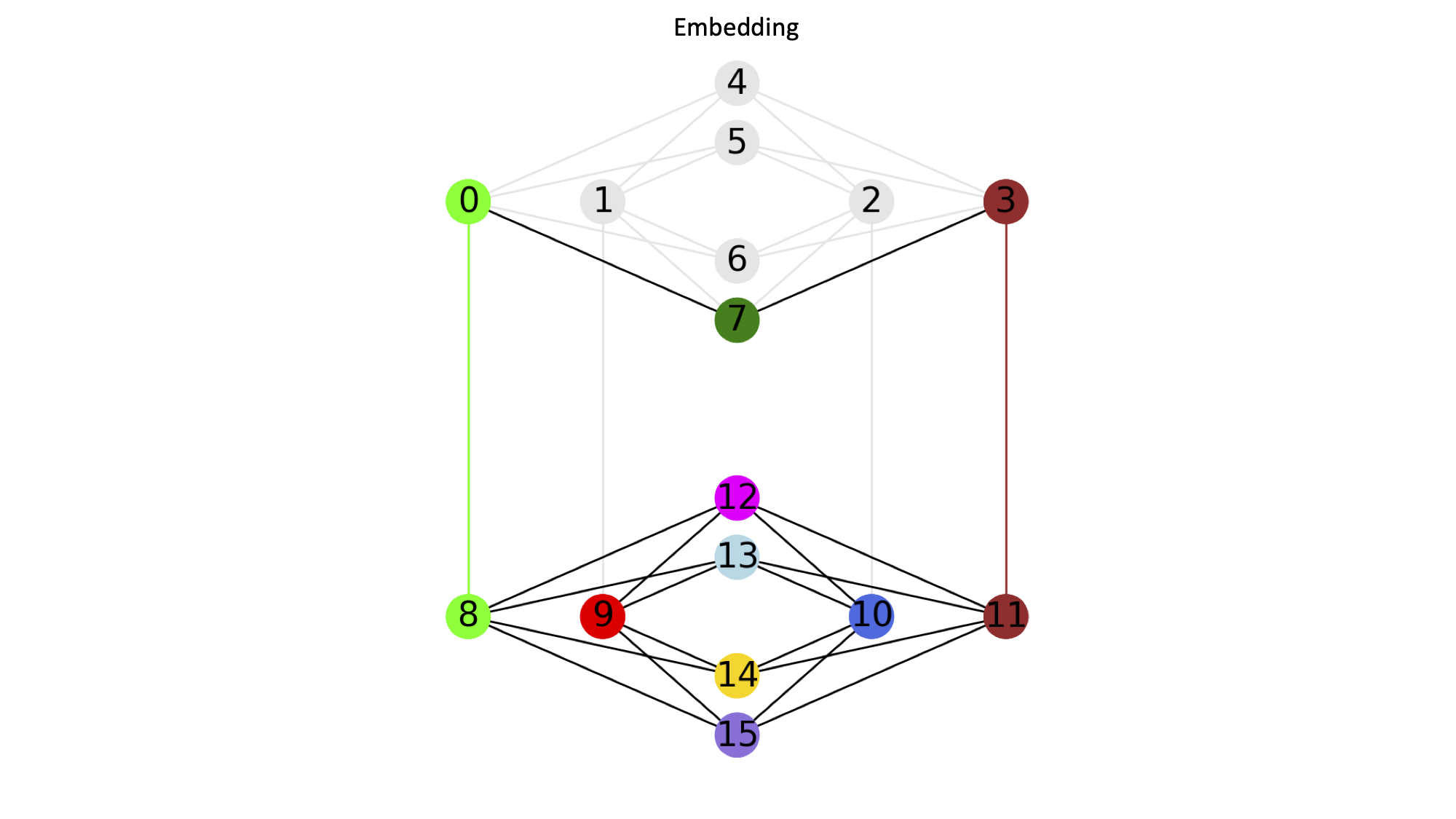
This example minor embeds a 3x3 grid graph onto a Chimera graph.
import networkx as nx
import dwave_networkx as dnx
import minorminer.layout as mml
grid_graph = nx.generators.lattice.grid_2d_graph(3, 3)
C = dnx.chimera_graph(2,1)
embedding = mml.find_embedding(grid_graph, C)
print(embedding)
# There are many possible outputs, and sometimes it might fail
# and return an empty list
# One run returned the following embedding:
{(0, 0): [13], (1, 0): [0, 8], (0, 1): [9], (1, 1): [12], (0, 2): [14], (1, 2): [10], (2, 0): [7], (2, 1): [11, 3], (2, 2): [15]}
Layout#
- class Layout(G, layout=None, dim=None, center=None, scale=None, pack_components=True, **kwargs)[source]#
Class that stores (or computes) coordinates in dimension
dimfor each node in graphG.- Parameters:
G (NetworkX Graph/edges data structure (dict, list, ...)) – The graph to compute the layout for or a NetworkX supported data structure for edges (see
nx.convert.to_networkx_graph()for details).layout (dict/function, optional, default=None) –
If a dict, this specifies a pre-computed layout for
G.If a function, this should be in the form of
layout(G, **kwargs), in whichdim,center, andscaleare passed in as kwargs and the return value is a dict representing a layout ofG.If None,
nx.random_layout(G, **kwargs)()is called.dim (int, optional, default=None) – The desired dimension of the computed layout, \(R^{\dim}\). If None,
dimis set as the dimension oflayout.center (tuple, optional, default=None) – The desired center point of the computed layout. If None,
centeris set as the center oflayout.scale (float, optional, default=None) – The desired scale of the computed layout; i.e. the layout is in \([center - scale, center + scale]^d\) space. If None,
scaleis set to be the scale oflayout.pack_components (bool, optional, default=True) – If True, and if the graph contains multiple components and
dimis None or 2, the components will be laid out individually and packed into a rectangle.**kwargs (dict) – Keyword arguments are passed to
layoutif it is a function.
Functions for Creating Layouts#
- p_norm(G, p=2, starting_layout=None, G_distances=None, dim=None, center=None, scale=None, **kwargs)[source]#
Embeds graph
Gin \(R^d\) with the p-norm and minimizes a Kamada-Kawai-esque objective function to achieve an embedding with low distortion.This computes a
LayoutforGwhere the graph distance and the p-distance are very close to each other.By default,
p_norm()is used to compute the layout for source graphs whenminorminer.layout.find_embedding()is called.- Parameters:
G (NetworkX Graph) – The graph to compute the layout for.
p (int, optional, default=2) – The order of the p-norm to use as a metric.
starting_layout (dict, optional, default=None) – A mapping from the vertices of
Gto points in \(R^d\). If None,nx.spectral_layout()is used if possible. Otherwise,nx.random_layout()is used.G_distances (dict, optional, default=None) – A dictionary of dictionaries representing distances from every vertex in
Gto every other vertex inG. If None, it is computed.dim (int, optional, default=None) – The desired dimension of the returned layout, \(R^{\dim}\). If None, the dimension of
centeris used. Ifcenteris None,dimis set to 2.center (tuple, optional, default=None) – The desired center point of the returned layout. If None,
centeris set as the origin in \(R^{\dim}\) space.scale (float, optional, default=None) – The desired scale of the returned layout; i.e. the layout is in \([center - scale, center + scale]^d\) space. If None, no scale is set.
- Returns:
Layout.layout, a mapping from vertices ofG(keys) to points in \(R^d\) (values).- Return type:
Examples
This example creates a
Layoutobject for a hexagonal lattice graph, with coordinates computed usingp_norm().>>> import networkx as nx >>> import minorminer.layout as mml ... >>> G = nx.hexagonal_lattice_graph(2,2) >>> layout = mml.Layout(G, mml.p_norm, center=(1,1))
layoutmay be passed in directly tominorminer.layout.find_embedding().Alternatively,
p_norm()may be passed in instead.This next example finds an embedding of a hexagonal lattice graph on a Chimera graph, in which the layouts of both the source and target graphs are computed using p_norm.
>>> import networkx as nx >>> import dwave_networkx as dnx >>> import minorminer.layout as mml ... >>> G = nx.hexagonal_lattice_graph(2,2) >>> C = dnx.chimera_graph(2,2) >>> embedding = mml.find_embedding(G, ... C, ... layout=(mml.p_norm, mml.p_norm), ... center=(1,1))
- dnx_layout(G, dim=None, center=None, scale=None, **kwargs)[source]#
The Chimera or Pegasus or Zephyr layout from dwave_networkx centered at the origin with
scaleas a function of the number of rows or columns. Note: As per the implementation of dnx.*_layout, if \(dim>2\), coordinates beyond the second are 0.By default,
dnx_layout()is used to compute the layout for target Chimera or Pegasus or Zephyr graphs whenminorminer.layout.find_embedding()is called.- Parameters:
G (NetworkX Graph) – The graph to compute the layout for.
dim (int, optional, default=None) – The desired dimension of the returned layout, \(R^{\dim}\). If None, the dimension of
centeris used. Ifcenteris None,dimis set to 2.center (tuple, optional, default=None) – The desired center point of the returned layout. If None, it is set as the origin in \(R^{\dim}\) space.
scale (float, optional, default=None) – The desired scale of the returned layout; i.e. the layout is in \([center - scale, center + scale]^d\) space. If None, it is set as \(max(n, m)/2\), where n, m are the number of columns, rows respectively in
G.
- Returns:
Layout.layout, a mapping from vertices ofG(keys) to points in \([-scale, scale]x[-scale, scale]x{0}^{d-2}+center\) (values).- Return type:
Examples
This example creates a
Layoutobject for a Pegasus graph, with coordinates computed usingdnx_layout().>>> import networkx as nx >>> import minorminer.layout as mml ... >>> P = dnx.pegasus_graph(4) >>> layout = mml.Layout(P, mml.dnx_layout, center=(1,1), scale=2)
Placement#
- class Placement(S_layout, T_layout, placement=None, scale_ratio=None, **kwargs)[source]#
Class that stores (or computes) a mapping of source nodes to collections of target nodes without any constraints. In mathematical terms, map V(S) to \(2^{V(T)}\).
- Parameters:
S_layout (
Layout) – A layout for S; i.e. a map from S to \(R^d\).T_layout (
Layout) – A layout for T; i.e. a map from T to \(R^d\).placement (dict/function, optional, default=None) –
If a dict, this specifies a pre-computed placement for S in T.
If a function, the function is called on
S_layoutandT_layout,placement(S_layout, T_layout), and should return a dict representing a placement of S in T.If None, a random placement of S in T is selected.
scale_ratio (float, optional, default=None) – If None,
S_layoutis not scaled. Otherwise,S_layoutis scaled toscale_ratio*T_layout.scale.**kwargs (dict) – Keyword arguments are passed to
placementif it is a function.
Functions for Creating Placements#
- intersection(S_layout, T_layout, **kwargs)[source]#
Map each vertex of S to its nearest row/column intersection qubit in T (T must be a D-Wave hardware graph). Note: This will modify
S_layout.- Parameters:
- Returns:
A mapping from vertices of S (keys) to vertices of T (values).
- Return type:
Examples
This example creates a
Placementobject that stores a mapping computed withintersection(), in which the nodes from a source hexagonal lattice graph are mapped to a target Chimera graph.>>> import networkx as nx >>> import dwave_networkx as dnx >>> import minorminer.layout as mml ... >>> G = nx.hexagonal_lattice_graph(2,2) >>> G_layout = mml.Layout(G, mml.p_norm) >>> C = dnx.chimera_graph(2,2) >>> C_layout = mml.Layout(C, mml.dnx_layout) >>> placement = mml.Placement(G_layout, C_layout, placement=mml.intersection)
placementmay be passed in directly tominorminer.layout.find_embedding().Alternatively,
intersection()may be passed in instead, as shown in the example below.>>> import networkx as nx >>> import dwave_networkx as dnx >>> import minorminer.layout as mml ... >>> G = nx.hexagonal_lattice_graph(2,2) >>> C = dnx.chimera_graph(2,2) >>> embedding = mml.find_embedding(G, ... C, ... placement=mml.intersection)
- closest(S_layout, T_layout, subset_size=(1, 1), num_neighbors=1, **kwargs)[source]#
Maps vertices of S to the closest vertices of T as given by
S_layoutandT_layout. i.e. For each vertex u inS_layoutand each vertex v inT_layout, map u to the v with minimum Euclidean distance \((||u - v||_2)\).By default,
closest()is used to compute the placement of an embedding whenminorminer.layout.find_embedding()is called.- Parameters:
S_layout (
Layout) – A layout for S; i.e. a map from S to \(R^d\).T_layout (
Layout) – A layout for T; i.e. a map from T to \(R^d\).subset_size (tuple, optional, default=(1, 1)) – A lower (subset_size[0]) and upper (subset_size[1]) bound on the size of subsets of T that will be considered when mapping vertices of S.
num_neighbors (int, optional, default=1) – The number of closest neighbors to query from the KDTree–the neighbor with minimum overlap is chosen. Increasing this reduces overlap, but increases runtime.
- Returns:
A mapping from vertices of S (keys) to subsets of vertices of T (values).
- Return type:
Examples
This example creates a
Placementobject that stores a mapping computed withclosest(), in which the nodes from a source hexagonal lattice graph are mapped to a target Chimera graph.>>> import networkx as nx >>> import dwave_networkx as dnx >>> import minorminer.layout as mml ... >>> G = nx.hexagonal_lattice_graph(2,2) >>> G_layout = mml.Layout(G, mml.p_norm) >>> C = dnx.chimera_graph(2,2) >>> C_layout = mml.Layout(C, mml.dnx_layout) >>> placement = mml.Placement(G_layout, C_layout, placement=mml.closest)